FDF Project
Main menu
- HOME
- Separador 1
- Product Information
- Separador 2
- Related Bibliography
- Separador 3
- FDF Kit Orders
- Separador 4
- Contact Us / Partners
It is well known that forensic related DNA is isolated from different sources, as blood stains placed over FTA paper, or buccal swabs, or biological stains founded in crime scene analysis.
The current DNA extraction methods from biological samples are performed using either protocols based in organic extractions or insoluble resins, or more recently through silica layers packed in columns or paramagnetics beads, able to retain DNA molecules, which once separated from contaminants and other components of the original sample, are then eluted from the active surface using high strong saline solutions,
But what about using sample DNA never touched or retained, only separated from other biomolecules or low molecular weight components?
Simple answer:
Fingerprint DNA Finder™: A New Approach
Nexttec's DNA extraction system is based on a reversal of the well-
FDF is able to obtain DNA from fingerprints (epidermal cells + cell-
Biomolecules and low molecular weight compounds are retained by sorbent.
DNA passes during a short, one-
Worried about thresholds? A simple vacuum centrifugation step fix the problem!
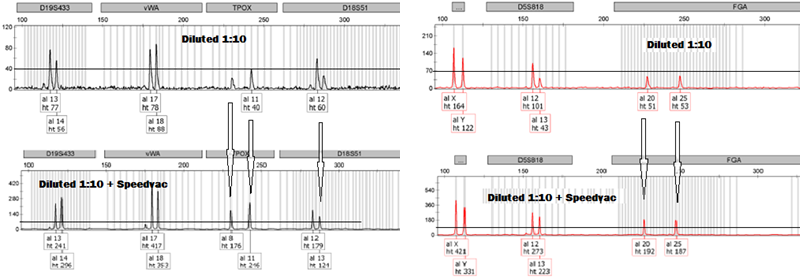
Partial electropherogram of an STR profile obtained from a fingerprint diluted 10 times, and concentrated to final 15 ul using a Speedvac centrifugation to medium heat. A concentration factor of approximately 4X is obtained.
